Highlights of the Month: February 2020
Published:
Key words for this month: Reformer; TMAP; libmolgrid;
Research Papers 🎓
Cheminformatics
Visualization of very large high-dimensional data sets as minimum spanning trees.
TMAP is a new data visualization method designed for visualizing large, high-dimensional data sets containing chemical structures and associated properties while preserving both global and local features.
Exploring chemical space using natural language processing methodologies for drug discovery Drug Discov Today | arXiv.
Text-based representations of chemicals and proteins can be thought of as unstructured languages codified by humans to describe domain-specific knowledge. Advances in natural language processing (NLP) methodologies in the processing of spoken languages accelerated the application of NLP to elucidate hidden knowledge in textual representations of these biochemical entities and then use it to construct models to predict molecular properties or to design novel molecules. This review outlines the impact made by these advances on drug discovery and aims to further the dialogue between medicinal chemists and computer scientists.
Towards reproducible computational drug discovery
This review explores the following topics: (1) the current state-of-the-art on reproducible research, (2) research documentation (e.g. electronic laboratory notebook, Jupyter notebook, etc.), (3) science of reproducible research (i.e. comparison and contrast with related concepts as replicability, reusability and reliability), (4) model development in computational drug discovery, (5) computational issues on model development and deployment, (6) use case scenarios for streamlining the computational drug discovery protocol.
A Deep Learning Approach to Antibiotic Discovery
A new machine-learning paper on antibiotic discovery has some pretty interesting results - a harbinger?https://t.co/v3nDF3D2Dl
— Derek Lowe (@Dereklowe) February 20, 2020
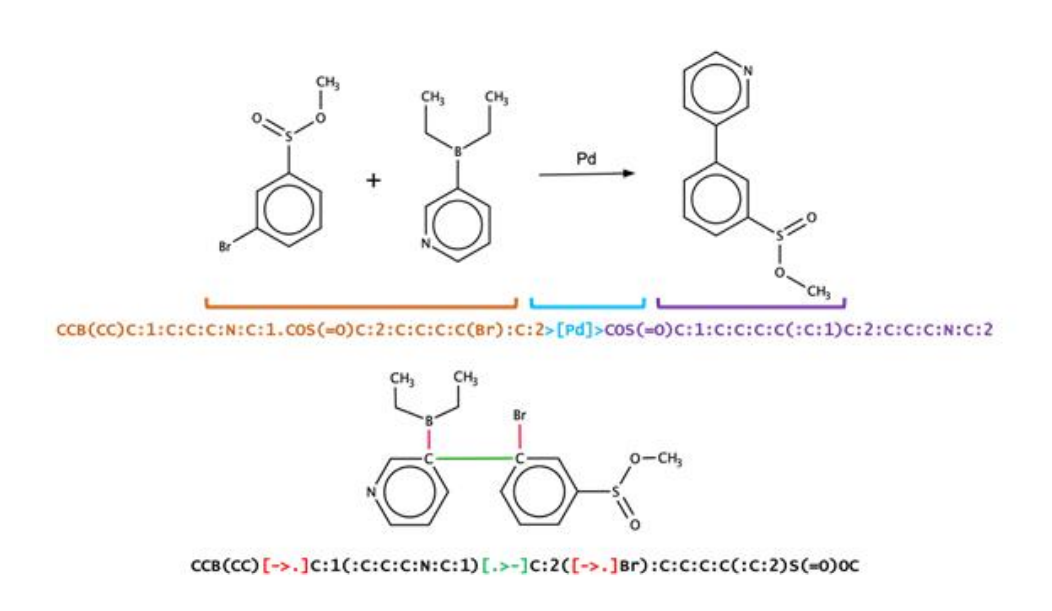
Discovery of Novel Chemical Reactions by Deep Generative Recurrent Neural Network
Condensed Graph of Reaction (CGR) encodes the structures of reactants and products into a single molecular graph (See Fig 1).
NLP
REALM : Retrieval-Augmented Language Model Pre-Training
To capture knowledge in a more modular and interpretable way, we augment language model pretraining with a latent knowledge retriever, which allows the model to retrieve and attend over documents from a large corpus such as Wikipedia, used during pre-training, fine-tuning and inference. For the first time, we show how to pre-train such a knowledge retriever in an unsupervised manner, using masked language modeling as the learning signal and backpropagating through a retrieval step that considers millions of documents.
Compressive Transformers for Long-Range Sequence Modelling
A new model and dataset for long-range memory.
ImageBERT: CROSS-MODAL PRE-TRAINING WITH LARGE-SCALE WEAK-SUPERVISED IMAGE-TEXT DATA.
ImageBERT is a new vision-language pre-trained model for image-text joint embedding from Microsoft. The model is pre-trained on four tasks simultaneously: Masked Language Modeling (MLM), Masked Object Classification (MOC), Masked Region Feature Regression (MRFR), and Image Text Matching (ITM). Along with the pre-trained model, the team also collected a Large-scale weAk-supervised Image-Text (LAIT) dataset from Web. One interesting finding of this paper is that multi-stage pre-training strategy outperforms single-stage pre-training.
Exploring the Limits of Transfer Learning with a Unified Text-to-Text Transformer
Transfer learning, where a model is first pre-trained on a data-rich task before being fine-tuned on a downstream task, has emerged as a powerful technique in natural language processing (NLP). The effectiveness of transfer learning has given rise to a diversity of approaches, methodology, and practice. In this paper, we explore the landscape of transfer learning techniques for NLP by introducing a unified framework that converts every language problem into a text-to-text format.
New dataset
22.5 million poses of ligands docked into multiple similar binding pockets across the Protein Data Bank and
others
A Scientist’s Guide to Social Media
Software and Tools 💻
TMAP is a new data visualization method designed for visualizing large, high-dimensional data sets containing chemical structures and associated properties while preserving both global and local features.
Best practices for computer vision from Microsoft. The goal for this repository is to build a comprehensive set of tools and examples that leverage recent advances in Computer Vision algorithms, neural architectures, and operationalizing such systems. This project is drawn from existing state-of-the-art libraries and build additional utility around loading image data, optimizing and evaluating models, and scaling up to the cloud. The examples are provided as Jupyter notebooks and common utility functions. All examples use Pytorch as the underlying deep learning library.
Represent 3D molecues using multidimensional arrays of voxelized molecular data for grid-based machine learning modeling.
Articles and Blog Posts 📃
Transformer - Illustration and code.ipynb
Re-implement OpenAI’s GPT-2 in PyTorch using Hugging Face source code and try to explain all the magic that goes on inside the model.
Train a language model from scratch using Hugging Face’s transformers and tokenizers
FastHugs - Fastai-v2 + HuggingFace
Fine-tuning a test classification model with HuggingFace transformer and the new fastai v2 library.
Interactive filtering of predicted ligands
Interesting application of ipywidgets, RDKit and jupyter notebook.
JAX is Numpy on the CPU, GPU and TPU, with greate automatic differentiation for high performance machine learning research.
Rethinking the Chemical Reaction as a Graph: Imaginary Transition Structures and Beyond
Self-Supervised Learning with Image网. How to get started with self-supervised learning, including examples of how to run and analyze experiments.
Notable Mentions ✨
The Art of the Algorithm: Machine Learning in Environmental Health Research. In this podcast, Dr. Nicole Kleinstreuer talked about the promise and potential pitfalls of artificial intelligence as it relates to environmental health research. She is the acting director of the Interagency Center for the Evaluation of Alternative Toxicological Methods within the National Toxicology Program at NIEHS.
[Colab Pro]. 10 Dollars/month, it delivers better GPUs and longer runtimes.
Leave a Comment